Least-Squares Fitting
Introduction
Curve Fitting Toolbox™ software uses the method of least squares when fitting data. Fitting requires a parametric model that relates the response data to the predictor data with one or more coefficients. The result of the fitting process is an estimate of the model coefficients.
To obtain the coefficient estimates, the least-squares method minimizes the summed square of residuals. The residual for the ith data point ri is defined as the difference between the observed response value yi and the fitted response value ŷi, and is identified as the error associated with the data.
The summed square of residuals is given by
where n is the number of data points included in the fit and S is the sum of squares error estimate. The supported types of least-squares fitting include:
Linear least squares
Weighted linear least squares
Robust least squares
Nonlinear least squares
Error Distributions
When fitting data that contains random variations, there are two important assumptions that are usually made about the error:
The error exists only in the response data, and not in the predictor data.
The errors are random and follow a normal (Gaussian) distribution with zero mean and constant variance, σ2.
The second assumption is often expressed as
The errors are assumed to be normally distributed because the normal distribution often provides an adequate approximation to the distribution of many measured quantities. Although the least-squares fitting method does not assume normally distributed errors when calculating parameter estimates, the method works best for data that does not contain a large number of random errors with extreme values. The normal distribution is one of the probability distributions in which extreme random errors are uncommon. However, statistical results such as confidence and prediction bounds do require normally distributed errors for their validity.
If the mean of the errors is zero, then the errors are purely random. If the mean is not zero, then it might be that the model is not the right choice for your data, or the errors are not purely random and contain systematic errors.
A constant variance in the data implies that the “spread” of errors is constant. Data that has the same variance is sometimes said to be of equal quality.
The assumption that the random errors have constant variance is not implicit to weighted least-squares regression. Instead, it is assumed that the weights provided in the fitting procedure correctly indicate the differing levels of quality present in the data. The weights are then used to adjust the amount of influence each data point has on the estimates of the fitted coefficients to an appropriate level.
Linear Least Squares
Curve Fitting Toolbox software uses the linear least-squares method to fit a linear model to data. A linear model is defined as an equation that is linear in the coefficients. For example, polynomials are linear but Gaussians are not. To illustrate the linear least-squares fitting process, suppose you have n data points that can be modeled by a first-degree polynomial.
To solve this equation for the unknown coefficients p1 and p2, you write S as a system of n simultaneous linear equations in two unknowns. If n is greater than the number of unknowns, then the system of equations is overdetermined.
Because the least-squares fitting process minimizes the summed square of the residuals, the coefficients are determined by differentiating S with respect to each parameter, and setting the result equal to zero.
The estimates of the true parameters are usually represented by b. Substituting b1 and b2 for p1 and p2, the previous equations become
where the summations run from i = 1 to n. The normal equations are defined as
Solving for b1
Solving for b2 using the b1 value
As you can see, estimating the coefficients p1 and p2 requires only a few simple calculations. Extending this example to a higher degree polynomial is straightforward although a bit tedious. All that is required is an additional normal equation for each linear term added to the model.
In matrix form, linear models are given by the formula
y = Xβ + ε
where
For the first-degree polynomial, the n equations in two unknowns are expressed in terms of y, X, and β as
The least-squares solution to the problem is a vector b, which estimates the unknown vector of coefficients β. The normal equations are given by
(XTX)b = XTy
where XT is the transpose of the design matrix X. Solving for b,
b = (XTX)–1 XTy
Use the MATLAB® backslash operator (mldivide) to solve a system
of simultaneous linear equations for unknown coefficients. Because inverting
XTX can lead to
unacceptable rounding errors, the backslash operator uses
QR decomposition with pivoting, which is a very
stable algorithm numerically. Refer to Arithmetic (MATLAB) for more
information about the backslash operator and QR
decomposition.
You can plug b back into the model formula to get the predicted response values, ŷ.
ŷ = Xb = Hy
H = X(XTX)–1 XT
A hat (circumflex) over a letter denotes an estimate of a parameter or a prediction from a model. The projection matrix H is called the hat matrix, because it puts the hat on y.
The residuals are given by
r = y – ŷ = (1–H)y
Weighted Least Squares
It is usually assumed that the response data is of equal quality and, therefore, has constant variance. If this assumption is violated, your fit might be unduly influenced by data of poor quality. To improve the fit, you can use weighted least-squares regression where an additional scale factor (the weight) is included in the fitting process. Weighted least-squares regression minimizes the error estimate
where wi are the weights. The weights determine how much each response value influences the final parameter estimates. A high-quality data point influences the fit more than a low-quality data point. Weighting your data is recommended if the weights are known, or if there is justification that they follow a particular form.
The weights modify the expression for the parameter estimates b in the following way,
where W is given by the diagonal elements of the weight matrix w.
You can often determine whether the variances are not constant by fitting the data and plotting the residuals. In the plot shown below, the data contains replicate data of various quality and the fit is assumed to be correct. The poor quality data is revealed in the plot of residuals, which has a “funnel” shape where small predictor values yield a bigger scatter in the response values than large predictor values.
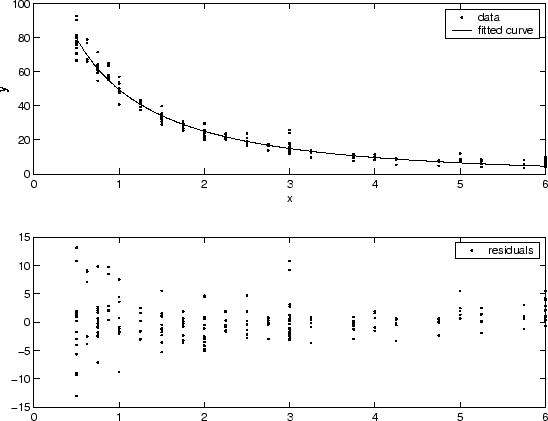
The weights you supply should transform the response variances to a constant value. If you know the variances of the measurement errors in your data, then the weights are given by
Or, if you only have estimates of the error variable for each data point, it usually suffices to use those estimates in place of the true variance. If you do not know the variances, it suffices to specify weights on a relative scale. Note that an overall variance term is estimated even when weights have been specified. In this instance, the weights define the relative weight to each point in the fit, but are not taken to specify the exact variance of each point.
For example, if each data point is the mean of several independent measurements, it might make sense to use those numbers of measurements as weights.
Robust Least Squares
It is usually assumed that the response errors follow a normal distribution, and that extreme values are rare. Still, extreme values called outliers do occur.
The main disadvantage of least-squares fitting is its sensitivity to outliers. Outliers have a large influence on the fit because squaring the residuals magnifies the effects of these extreme data points. To minimize the influence of outliers, you can fit your data using robust least-squares regression. The toolbox provides these two robust regression methods:
Least absolute residuals (LAR) — The LAR method finds a curve that minimizes the absolute difference of the residuals, rather than the squared differences. Therefore, extreme values have a lesser influence on the fit.
Bisquare weights — This method minimizes a weighted sum of squares, where the weight given to each data point depends on how far the point is from the fitted line. Points near the line get full weight. Points farther from the line get reduced weight. Points that are farther from the line than would be expected by random chance get zero weight.
For most cases, the bisquare weight method is preferred over LAR because it simultaneously seeks to find a curve that fits the bulk of the data using the usual least-squares approach, and it minimizes the effect of outliers.
Robust fitting with bisquare weights uses an iteratively reweighted least-squares algorithm, and follows this procedure:
Fit the model by weighted least squares.
Compute the adjusted residuals and standardize them. The adjusted residuals are given by
ri are the usual least-squares residuals and hi are leverages that adjust the residuals by reducing the weight of high-leverage data points, which have a large effect on the least-squares fit. The standardized adjusted residuals are given by
K is a tuning constant equal to 4.685, and s is the robust variance given by MAD/0.6745 where MAD is the median absolute deviation of the residuals.
Compute the robust weights as a function of u. The bisquare weights are given by
Note that if you supply your own regression weight vector, the final weight is the product of the robust weight and the regression weight.
If the fit converges, then you are done. Otherwise, perform the next iteration of the fitting procedure by returning to the first step.
The plot shown below compares a regular linear fit with a robust fit using bisquare weights. Notice that the robust fit follows the bulk of the data and is not strongly influenced by the outliers.
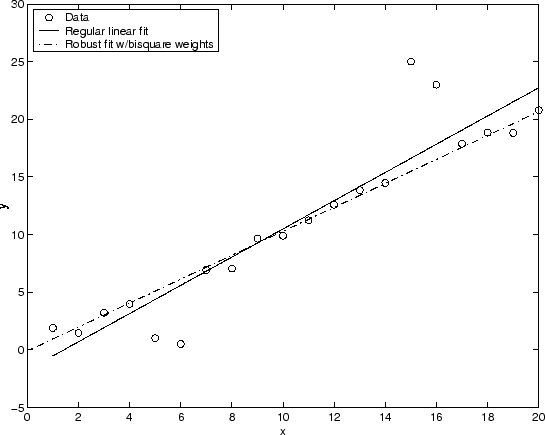
Instead of minimizing the effects of outliers by using robust regression, you can mark data points to be excluded from the fit. Refer to Remove Outliers for more information.
Nonlinear Least Squares
Curve Fitting Toolbox software uses the nonlinear least-squares formulation to fit a nonlinear model to data. A nonlinear model is defined as an equation that is nonlinear in the coefficients, or a combination of linear and nonlinear in the coefficients. For example, Gaussians, ratios of polynomials, and power functions are all nonlinear.
In matrix form, nonlinear models are given by the formula
y = f(X,β) + ε
where
y is an n-by-1 vector of responses.
f is a function of β and X.
β is a m-by-1 vector of coefficients.
X is the n-by-m design matrix for the model.
ε is an n-by-1 vector of errors.
Nonlinear models are more difficult to fit than linear models because the coefficients cannot be estimated using simple matrix techniques. Instead, an iterative approach is required that follows these steps:
Start with an initial estimate for each coefficient. For some nonlinear models, a heuristic approach is provided that produces reasonable starting values. For other models, random values on the interval [0,1] are provided.
Produce the fitted curve for the current set of coefficients. The fitted response value ŷ is given by
ŷ = f(X,b)
and involves the calculation of the Jacobian of f(X,b), which is defined as a matrix of partial derivatives taken with respect to the coefficients.
Adjust the coefficients and determine whether the fit improves. The direction and magnitude of the adjustment depend on the fitting algorithm. The toolbox provides these algorithms:
Trust-region — This is the default algorithm and must be used if you specify coefficient constraints. It can solve difficult nonlinear problems more efficiently than the other algorithms and it represents an improvement over the popular Levenberg-Marquardt algorithm.
Levenberg-Marquardt — This algorithm has been used for many years and has proved to work most of the time for a wide range of nonlinear models and starting values. If the trust-region algorithm does not produce a reasonable fit, and you do not have coefficient constraints, you should try the Levenberg-Marquardt algorithm.
Iterate the process by returning to step 2 until the fit reaches the specified convergence criteria.
You can use weights and robust fitting for nonlinear models, and the fitting process is modified accordingly.
Because of the nature of the approximation process, no algorithm is foolproof for all nonlinear models, data sets, and starting points. Therefore, if you do not achieve a reasonable fit using the default starting points, algorithm, and convergence criteria, you should experiment with different options. Refer to Specifying Fit Options and Optimized Starting Points for a description of how to modify the default options. Because nonlinear models can be particularly sensitive to the starting points, this should be the first fit option you modify.
Robust Fitting
This example shows how to compare the effects of excluding outliers and robust fitting. The example shows how to exclude outliers at an arbitrary distance greater than 1.5 standard deviations from the model. The steps then compare removing outliers with specifying a robust fit which gives lower weight to outliers.
Create a baseline sinusoidal signal:
xdata = (0:0.1:2*pi)'; y0 = sin(xdata);
Add noise to the signal with nonconstant variance.
% Response-dependent Gaussian noise gnoise = y0.*randn(size(y0)); % Salt-and-pepper noise spnoise = zeros(size(y0)); p = randperm(length(y0)); sppoints = p(1:round(length(p)/5)); spnoise(sppoints) = 5*sign(y0(sppoints)); ydata = y0 + gnoise + spnoise;
Fit the noisy data with a baseline sinusoidal model, and specify 3 output arguments to get fitting information including residuals.
f = fittype('a*sin(b*x)'); [fit1,gof,fitinfo] = fit(xdata,ydata,f,'StartPoint',[1 1]);
Examine the information in the fitinfo structure.
fitinfo
fitinfo = struct with fields:
numobs: 63
numparam: 2
residuals: [63x1 double]
Jacobian: [63x2 double]
exitflag: 3
firstorderopt: 0.0883
iterations: 5
funcCount: 18
cgiterations: 0
algorithm: 'trust-region-reflective'
stepsize: 4.1539e-04
message: 'Success, but fitting stopped because change in residuals less than tolerance (TolFun).'
Get the residuals from the fitinfo structure.
residuals = fitinfo.residuals;
Identify "outliers" as points at an arbitrary distance greater than 1.5 standard deviations from the baseline model, and refit the data with the outliers excluded.
I = abs( residuals) > 1.5 * std( residuals ); outliers = excludedata(xdata,ydata,'indices',I); fit2 = fit(xdata,ydata,f,'StartPoint',[1 1],... 'Exclude',outliers);
Compare the effect of excluding the outliers with the effect of giving them lower bisquare weight in a robust fit.
fit3 = fit(xdata,ydata,f,'StartPoint',[1 1],'Robust','on');
Plot the data, the outliers, and the results of the fits. Specify an informative legend.
plot(fit1,'r-',xdata,ydata,'k.',outliers,'m*') hold on plot(fit2,'c--') plot(fit3,'b:') xlim([0 2*pi]) legend( 'Data', 'Data excluded from second fit', 'Original fit',... 'Fit with points excluded', 'Robust fit' ) hold off

Plot the residuals for the two fits considering outliers:
figure plot(fit2,xdata,ydata,'co','residuals') hold on plot(fit3,xdata,ydata,'bx','residuals') hold off
